Independent Set Problem
In graph theory, an independent set, stable set, coclique or anticlique is a set of vertices in a graph, no two of which are adjacent. That is, it is a set \(S\) of vertices such that for every two vertices in \(S\), there is no edge connecting the two. Equivalently, each edge in the graph has at most one endpoint in \(S\). A set is independent if and only if it is a clique in the graph’s complement. The size of an independent set is the number of vertices it contains. Independent sets have also been called “internally stable sets”, of which “stable set” is a shortening. The independent set decision problem is \(NP\)-complete, and hence it is not believed that there is an efficient algorithm for solving it.
A maximal independent set is an independent set that is not a proper subset of any other independent set. A maximum independent set is an independent set of largest possible size for a given graph \(G\). This size is called the independence number of \(G\) and is usually denoted by \(\alpha (G)\). The optimization problem of finding such a set is called the maximum independent set problem. It is a strongly NP-hard problem. As such, it is unlikely that there exists an efficient algorithm for finding a maximum independent set of a graph. Every maximum independent set also is maximal, but the converse implication does not necessarily hold.
Finding maximal independent set
For a graph \(G(V,E)\), the integer programming model for finding the vertices that belong to the maximal independent set is as follows
\[ \max \sum_{i \in V} x_i \\\\ s.t. x_i + x_j \leq 1, \text{ \$\forall (i,j) \in E \$} \\\\ x \in \{0,1\}^{|V|} \]
SAT and \(3\)-SAT
The Boolean Satisfiability Problem or in other words SAT is the first problem that was shown to be NP-Complete. A given boolean expression, determining if there exists a truth assignment to its variables for which the expression is true is defined as the satisfiability problem.
When we’re discussing the SAT problem, it’s essential to know about the conjunctive normal form (CNF). A boolean expression is said to be in CNF form if it’s a conjunction of a set of clauses, where each clause is defined as a disjunction (logical OR) of literals. We can define a literal as a variable or a negation of a variable.
Every boolean expression can be expressed as a CNF by repeatedly using De Morgan’s Laws, the law of double negation and distributive laws under AND and OR:
\[C_1 \wedge C_2 \wedge ... \wedge C_n\]
where,
\[C_i = a_1^i \vee a_2^i \vee ... \vee a_k^i\]
The above CNF is a \(k\)-CNF, that is, it has a maximum of \(k\) literals in every clause. We pick each literal from a set of \(m\) variables: \(x_1, x_2,...,x_m, \neg x_1, \neg x_2,..., \neg x_m\)
3-SAT
In the 3-SAT problem, each clause has at most \(3\) literals. This problem is NP-Complete. The 3-SAT problem is part of the Karp’s \(21\) NP-complete problems and is used as the starting point to prove that the other problems are also NP-Complete. One example is the independent set problem.
Reduction of \(3\)-SAT to Independent Set Problem
\(G\) will have one vertex for each literal in a clause.
Connect the \(3\)-literals in a clause to form a triangle; the independent set will pick at most one vertex from each clause, which will correspond to the literal to be set to true.
Connect \(2\) vertices if they label complementary literals; this ensures that the literals corresponding to the independent set do not have a conflict.
Take \(k\) to be the number of clauses.
Example
If \(\phi = (x \vee y \vee z)(\bar{x}\vee\bar{y}\vee z)(x\vee\bar{y}\vee \bar{z})(\bar{z}\vee y \vee \bar{z})\). The graph corresponding to the given \(3\)-SAT instance is given below 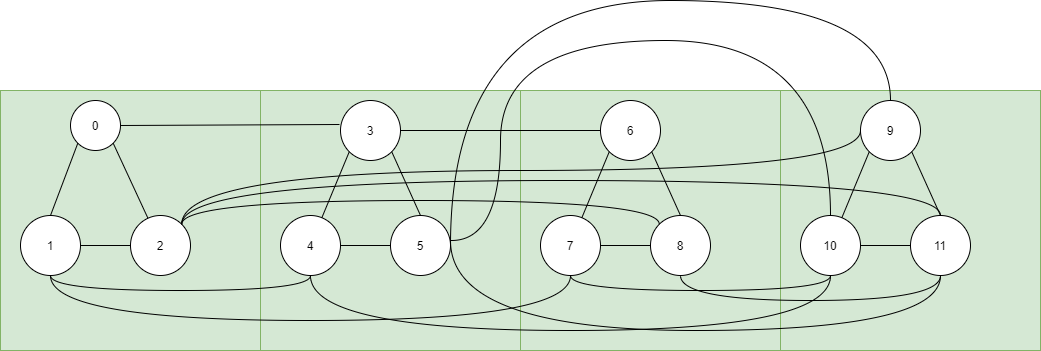
The corresponding incidence matrix of the graph can be found here.
The incidence matrix of graph \(G\) on \(n\) vertices and \(m\) edges is an \(n \times m\) matrix \(A = [a_{ij}]\) such that
\[ a_{ij} = \begin{cases} 1 \text{, if edge \$j \in E$ is incident on vertex \$i \in V \$} \\\\ 0 \text{, otherwise} \\\\ \end{cases} \]
The model for finding the maximal indepedent set using the incidence matrix is given by:
\[ \max \sum_{i \in V} x_i \\\\ s.t. \sum_{i \in V} a_{ij} x_i \leq 1, \text{ \$\forall j \in E \$} \\\\ x \in \{0,1\}^{|V|} \]
The code for solving the \(3\)-SAT problem by converting it into a graph (represented by an incidence matrix) using the reduction process mentioned above is given below
Code using Gurobipy
import gurobipy as gp
from gurobipy import GRB, quicksum
def inc_mat(filepath):
import re
with open(filepath,'r') as f:
return [[int(i) for i in re.findall(r'-?\d+', line)] for line in f]
def max_ind_set(inc_mat):
n_vertices, n_edges = len(inc_mat), len(inc_mat[0])
isp = gp.Model("ISP")
x = isp.addVars(range(n_vertices), vtype = GRB.BINARY, name ="x")
isp.addConstrs((quicksum([inc_mat[i][j]*x[i] for i in range(n_vertices)]) <= 1
for j in range(n_edges)))
obj = quicksum([x[i] for i in range(n_vertices)])
isp.setObjective(obj, GRB.MAXIMIZE)
isp.optimize()
if isp.status == GRB.OPTIMAL:
return [int(x[i].x) for i in range(n_vertices)]
else:
return None
FILEPATH = "incidence_matrix.txt"
print(max_ind_set(inc_mat(FILEPATH)))The output is \([1, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0]\) corresponding to choosing \(x\) in the first clause, \(\bar{y}\) in the second clause, \(x\) in the third clause, \(\bar{z}\) in the fourth clause.
Maximal Clique
In computer science, the clique problem is the computational problem of finding cliques (subsets of vertices, all adjacent to each other, also called complete subgraphs) in a graph. Common formulations of the clique problem include finding a maximum clique (a clique with the largest possible number of vertices), finding a maximum weight clique in a weighted graph, listing all maximal cliques (cliques that cannot be enlarged), and solving the decision problem of testing whether a graph contains a clique larger than a given size.
A maximal clique is a set of vertices that induces a complete subgraph, and that is not a subset of the vertices of any larger complete subgraph. That is, it is a set \(S\) such that every pair of vertices in \(S\) is connected by an edge and every vertex not in \(S\) is missing an edge to at least one vertex in \(S\). A graph may have many maximal cliques, of varying sizes; finding the largest of these is the maximum clique problem.
The independent set problem and the clique problem are complementary: a clique in \(G\) is an independent set in the complement graph of \(G\) and vice versa. If \(S\) is a maximal independent set in some graph, it is a maximal clique or maximal complete subgraph in the complementary graph.
Therefore, we can reuse the code for determining the maximal independent set of a graph. All we needs is an additional function that returns the incidence matrix of the complementary graph given the original incidence matrix. The code for solving the maximal clique problem given an incidence matrix is given below.
Example
Find the maximal clique in the graph below:
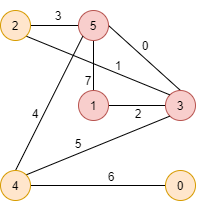
The incidence matrix for the graph is
\[ \begin{bmatrix} 0& 0 & 0 & 0 & 0 & 0 & 1 & 0\\\\ 0& 0 & 1 & 0 & 0 & 0 & 0 & 1\\\\ 0& 1 & 0 & 1 & 0 & 0 & 0 & 0\\\\ 1& 1 & 1 & 0 & 0 & 1 & 0 & 0\\\\ 0& 0 & 0 & 0 & 1 & 1 & 1 & 0\\\\ 1& 0 & 0 & 1 & 1 & 0 & 0 & 1\\\\ \end{bmatrix} \]
Code using Gurobipy
import gurobipy as gp
from gurobipy import GRB, quicksum
from itertools import combinations
def comp_inc_mat(inc_mat):
def mat_t(mat):
n_r, n_c = len(mat), len(mat[0])
return [[mat[j][i] for j in range(n_r)] for i in range(n_c)]
n_v, inc_mat_t, c_inc_mat = len(inc_mat), mat_t(inc_mat), []
for j, k in combinations(range(n_v),2):
r = [0 for _ in range(n_v)]
r[j], r[k] = 1, 1
if r not in inc_mat_t:
c_inc_mat.append(r)
return mat_t(c_inc_mat)
def max_ind_set(inc_mat):
n_vertices, n_edges = len(inc_mat), len(inc_mat[0])
isp = gp.Model("ISP")
x = isp.addVars(range(n_vertices), vtype = GRB.BINARY, name ="x")
isp.addConstrs((quicksum([inc_mat[i][j]*x[i] for i in range(n_vertices)]) <= 1
for j in range(n_edges)))
obj = quicksum([x[i] for i in range(n_vertices)])
isp.setObjective(obj, GRB.MAXIMIZE)
isp.optimize()
if isp.status == GRB.OPTIMAL:
return [int(x[i].x) for i in range(n_vertices)]
else:
return None
inc_mat_1 = [[0, 0 , 0 , 0 , 0 , 0 , 1 , 0],
[0, 0 , 1 , 0 , 0 , 0 , 0 , 1],
[0, 1 , 0 , 1 , 0 , 0 , 0 , 0],
[1, 1 , 1 , 0 , 0 , 1 , 0 , 0],
[0, 0 , 0 , 0 , 1 , 1 , 1 , 0],
[1, 0 , 0 , 1 , 1 , 0 , 0 , 1]]
print(max_ind_set(comp_inc_mat(inc_mat_1)))The output is \([0, 1, 0, 1, 0, 1]\), the set of vertices \(\{1,3,5\}\) in the maximal independent set in the complementary graph which is the same as the set of vertices in the maximal clique in the original graph.